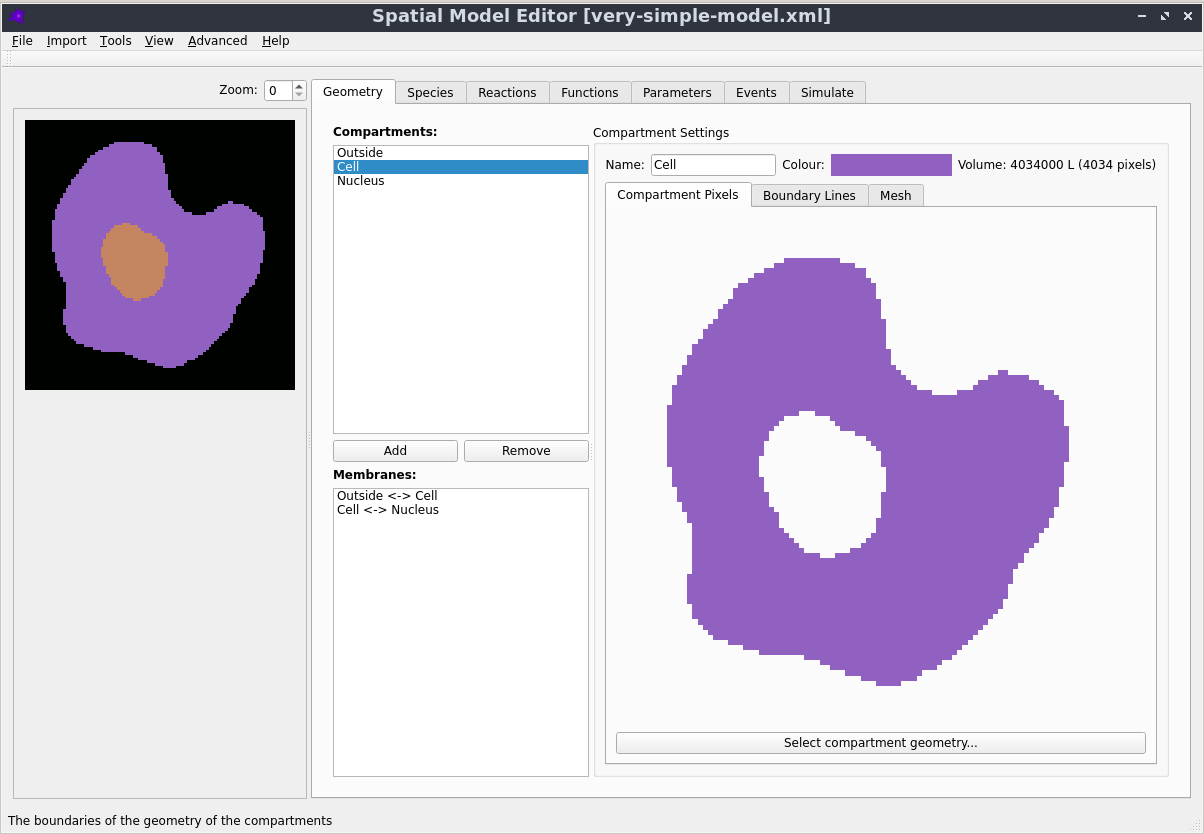
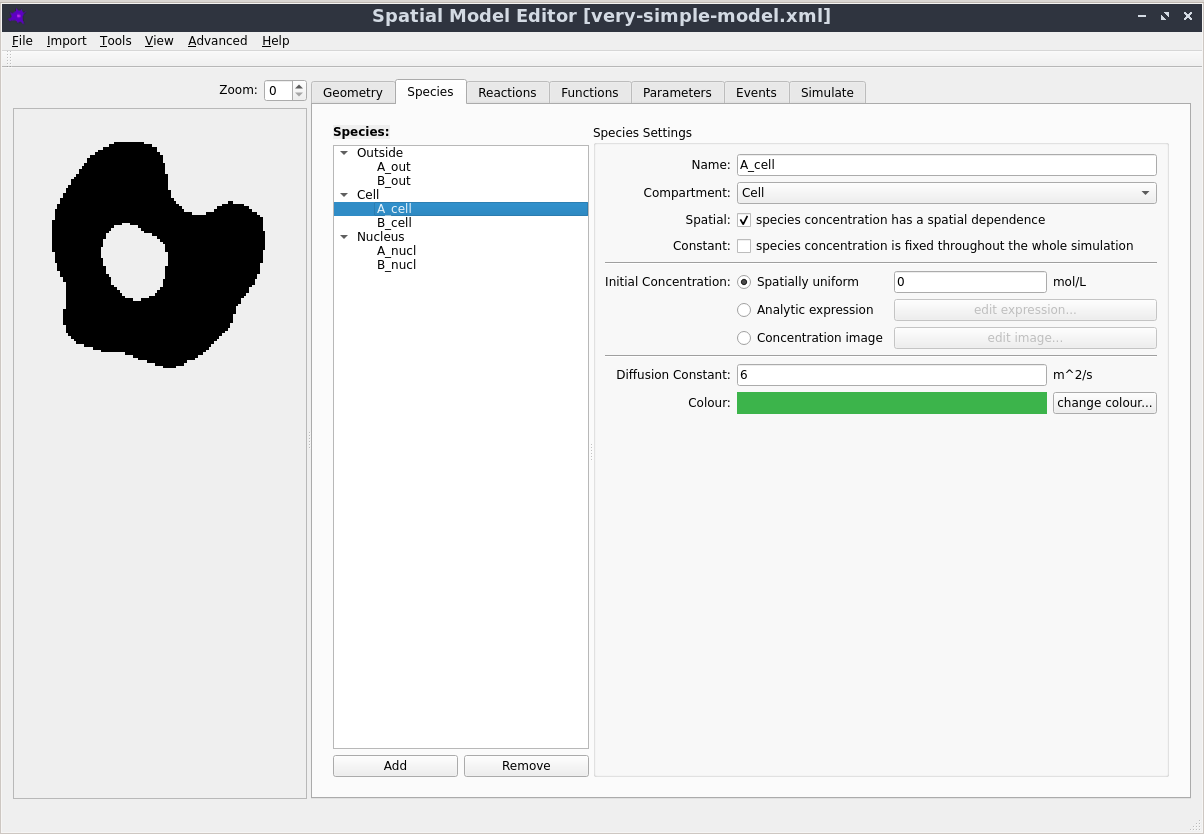
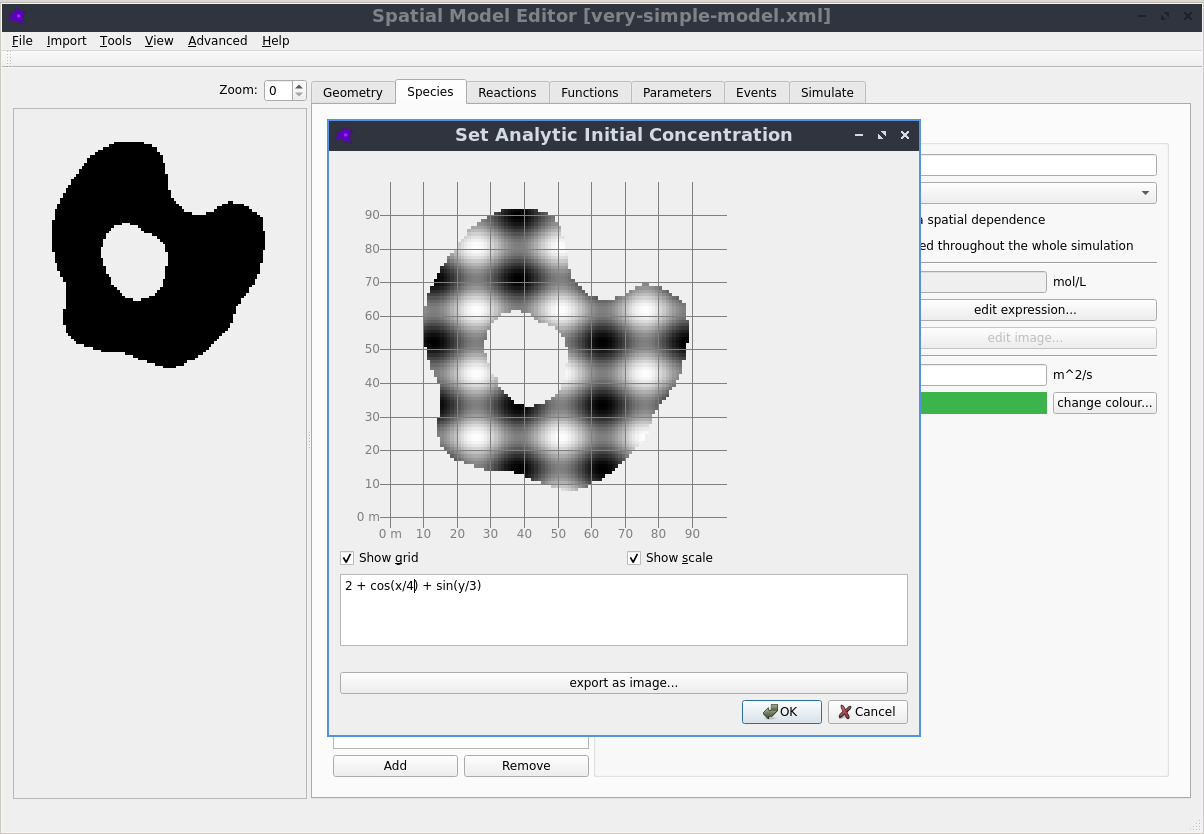
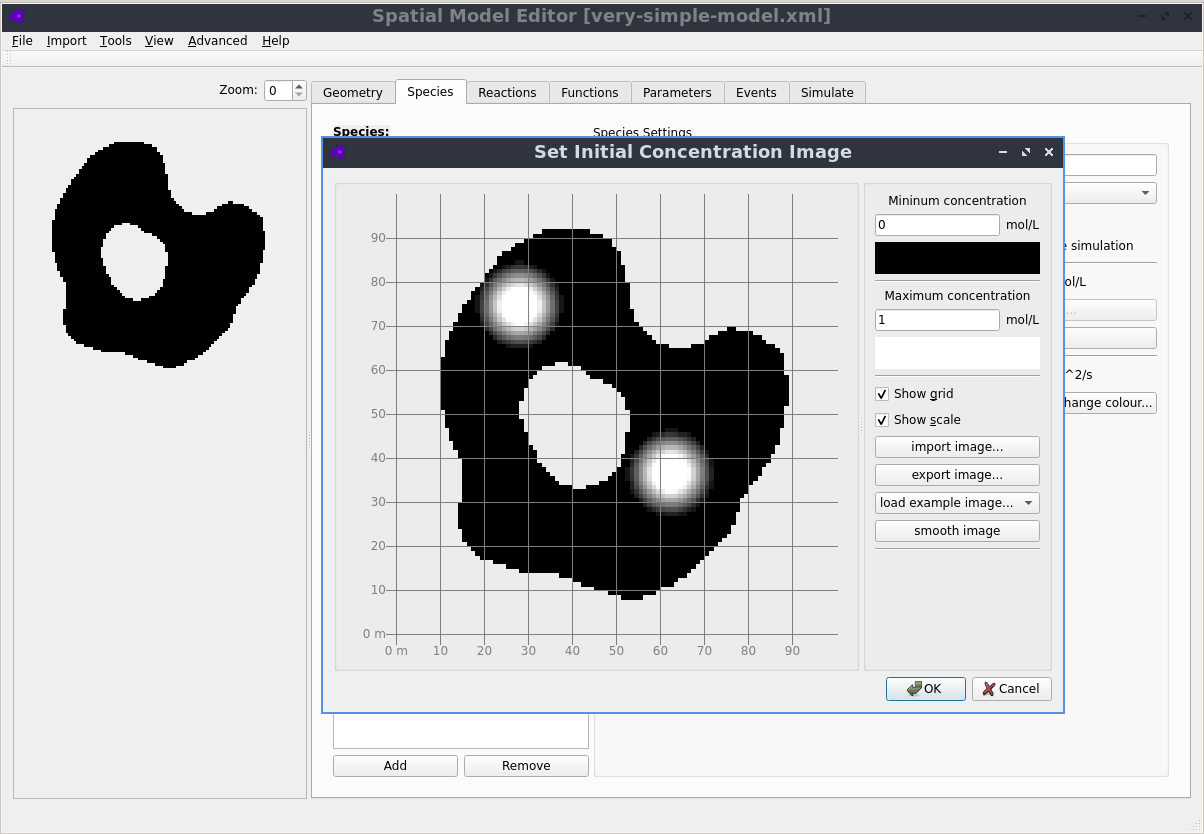
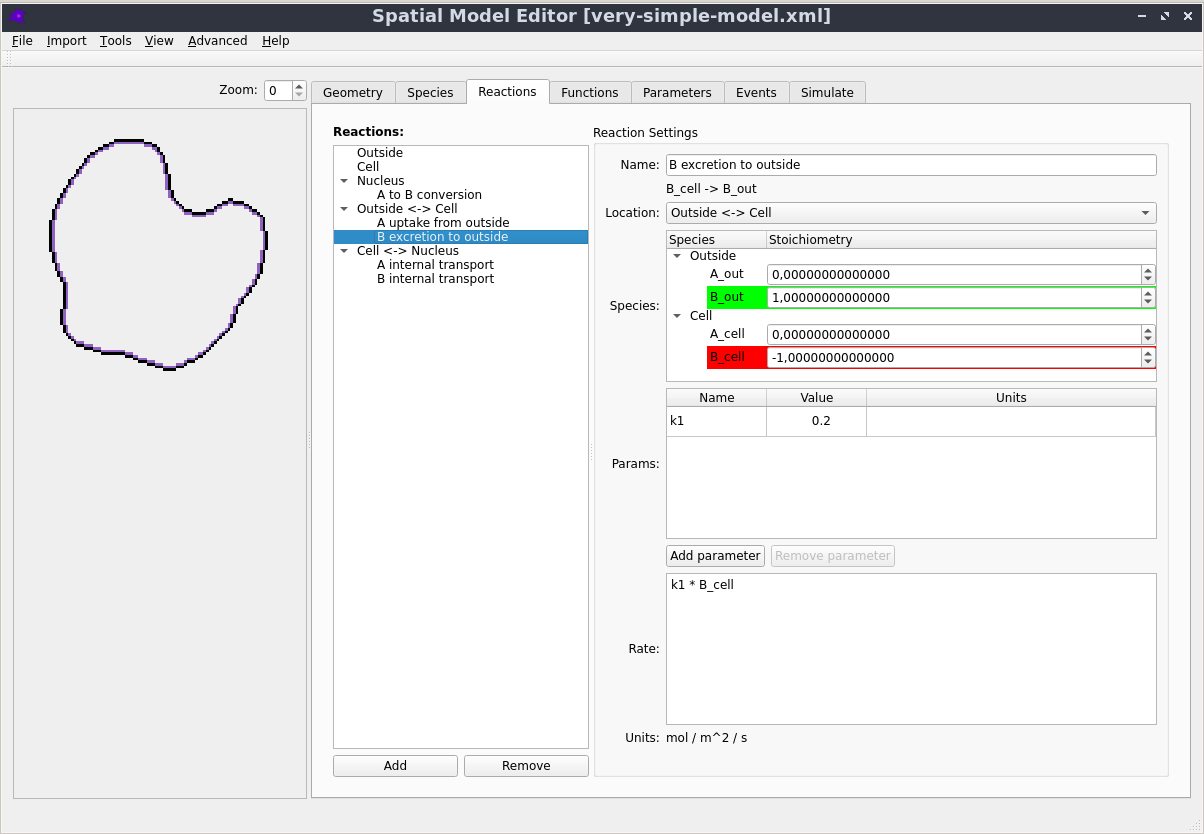
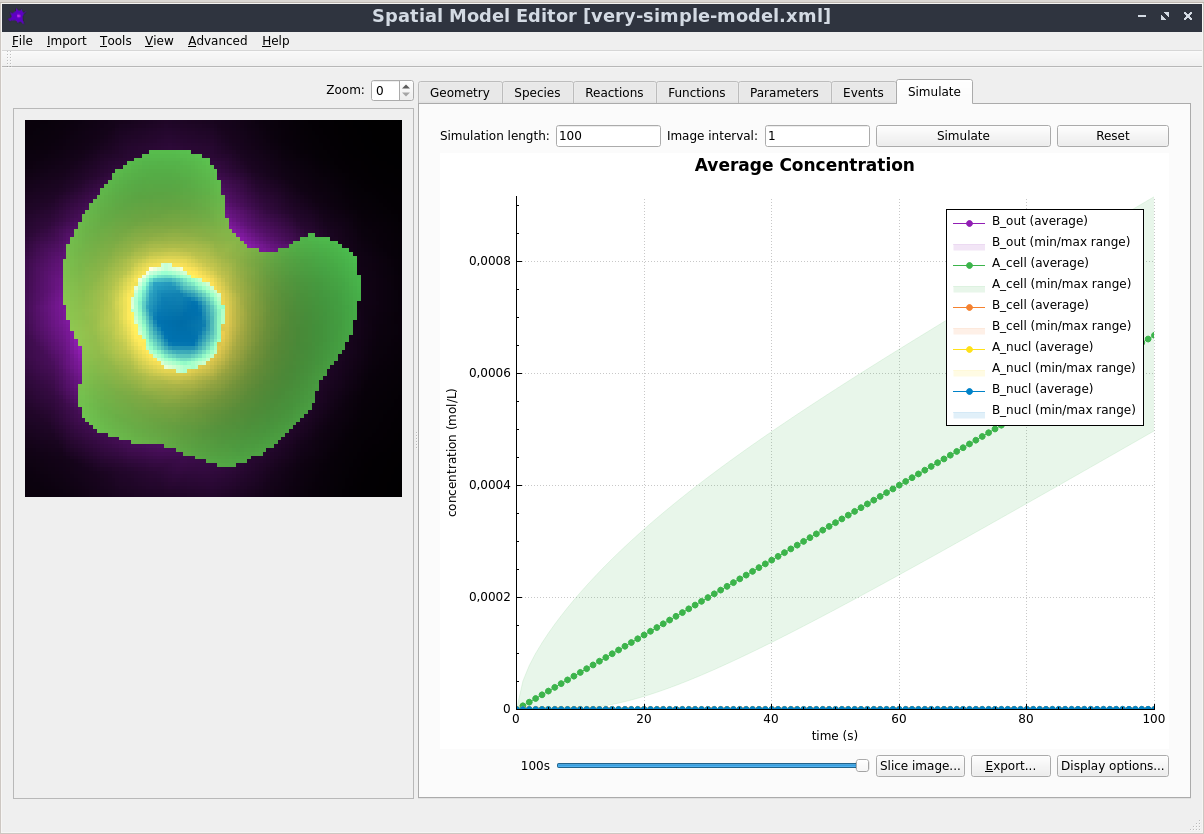
Graphical User Interface
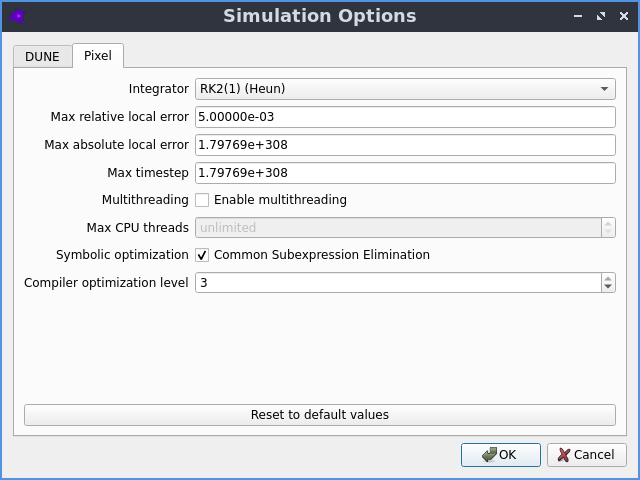
Creating, editing and simulating a spatial model is all done using a user friendly Graphical User Interface. See the Getting Started section in the documentation to learn more.
DuneCopasi FEM Solver
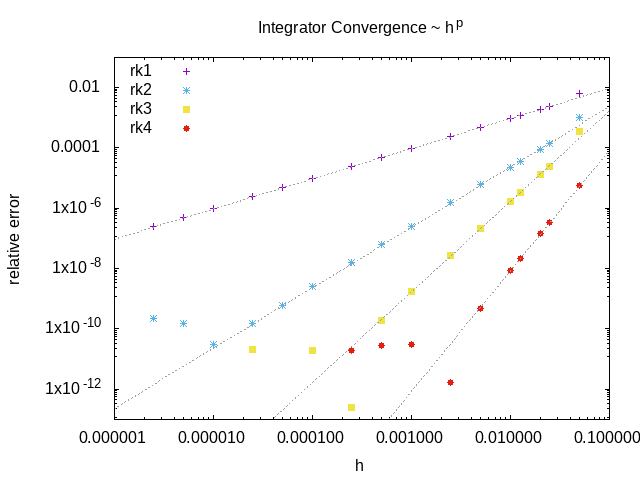
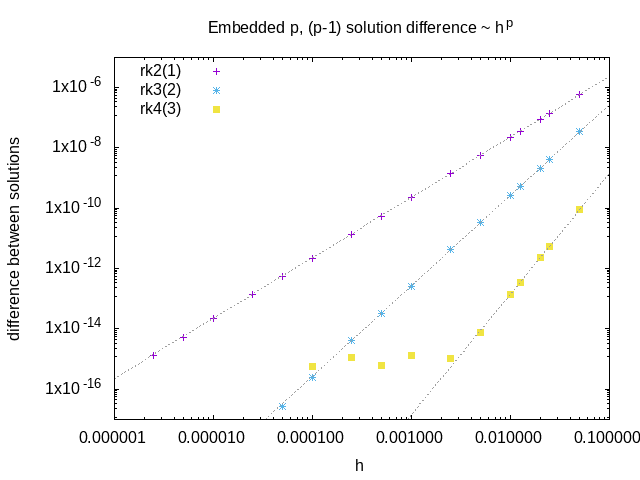
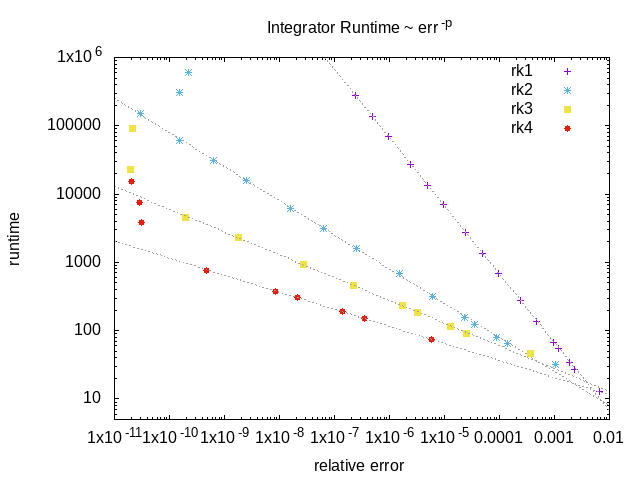
The dune-copasi solver is used to simulate models. This solves the PDE on a tetrahedral mesh using finite element discretization methods. See the simulator documentation for more information on the available options when using it in the GUI. For more technical information about the DuneCopasi solver itself, see the dune-copasi website.
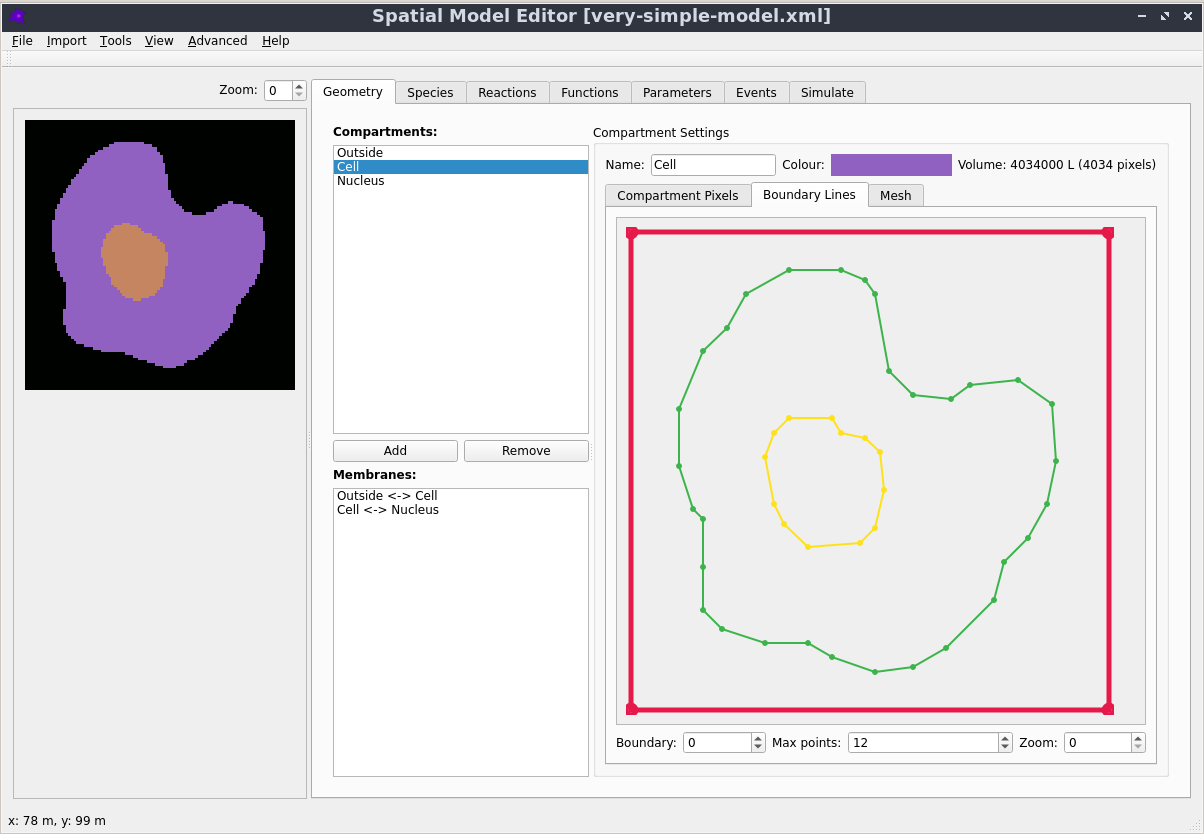
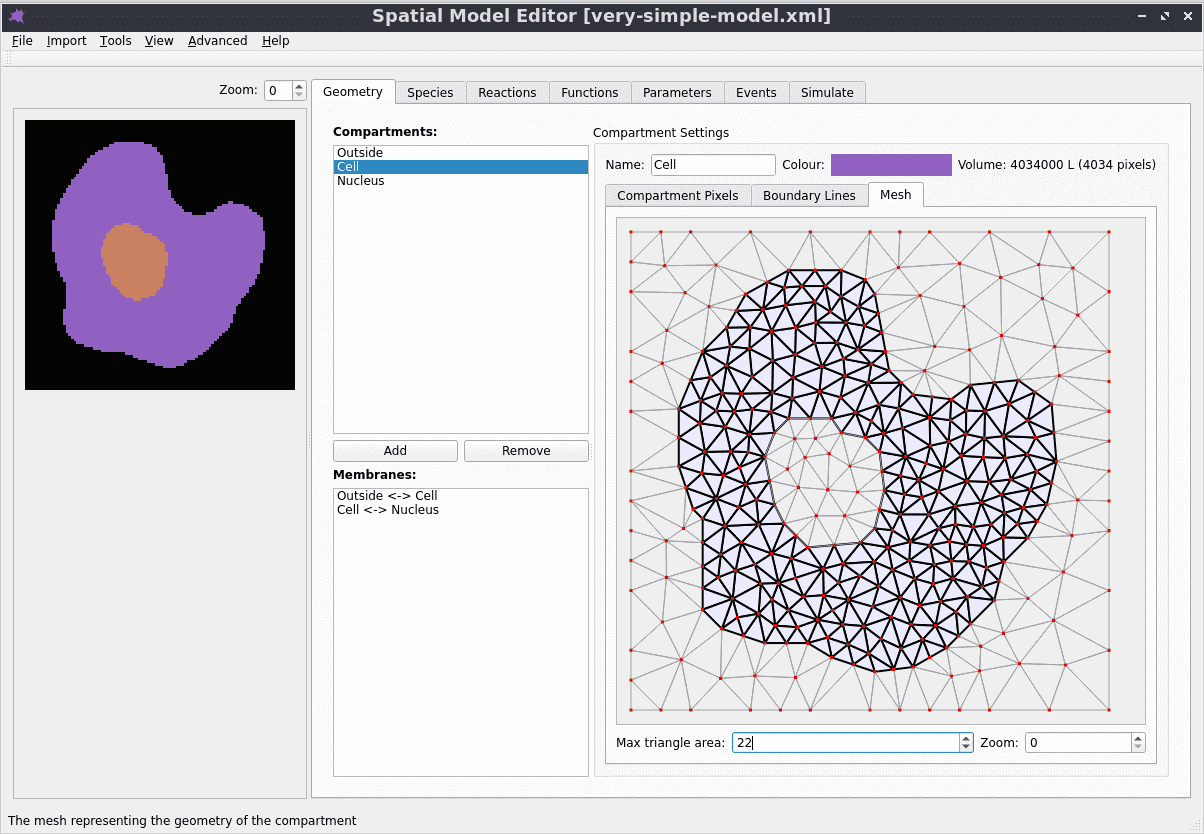
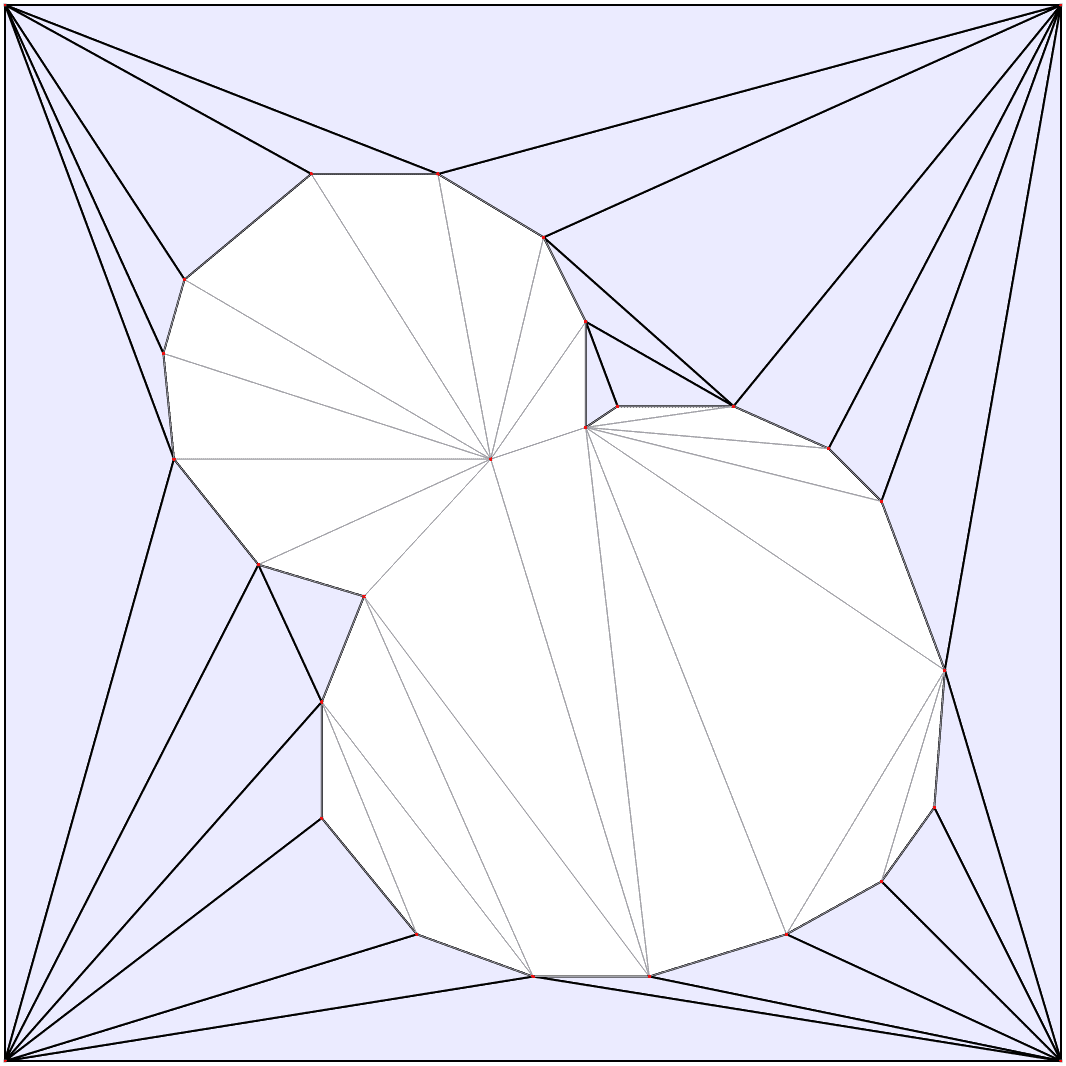
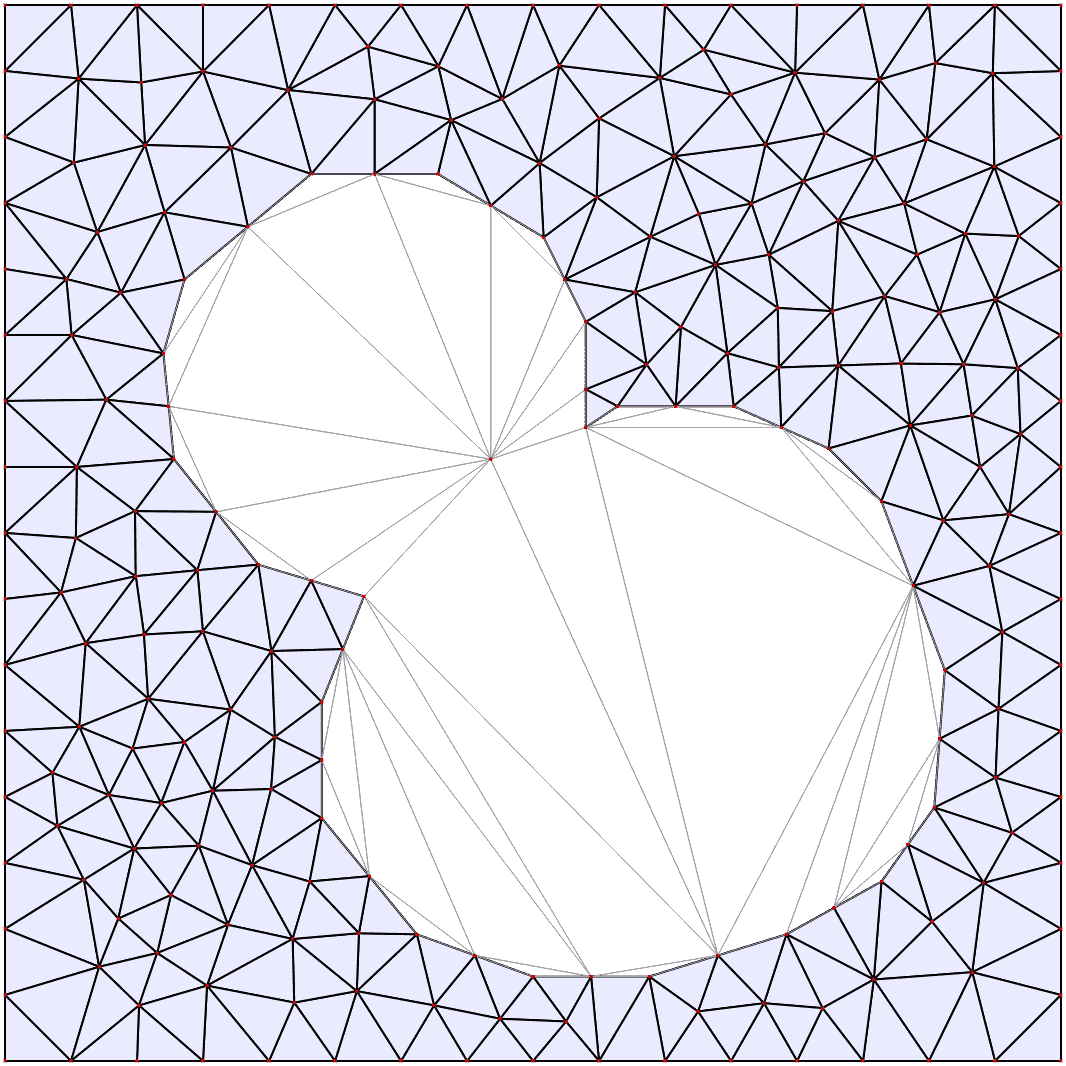
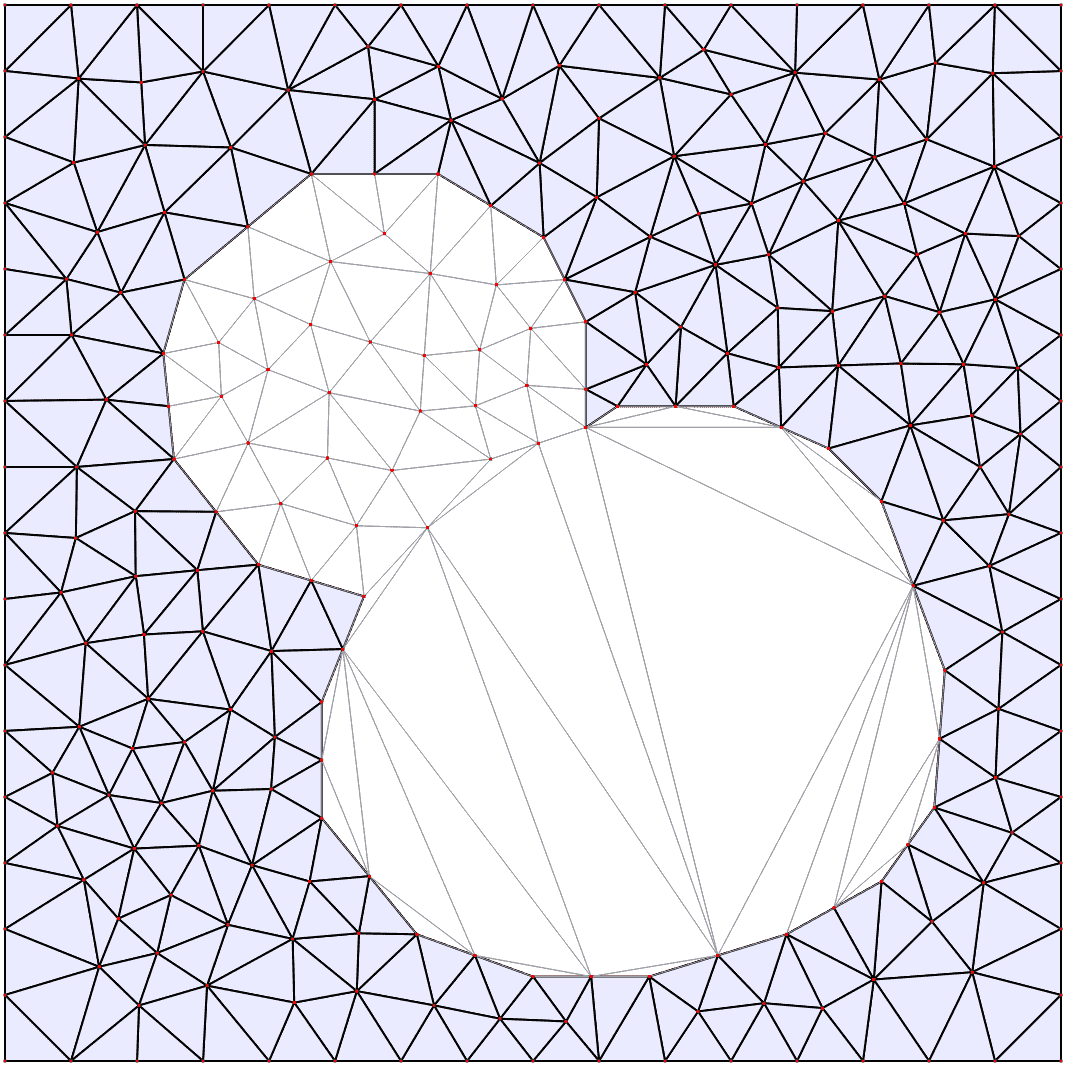
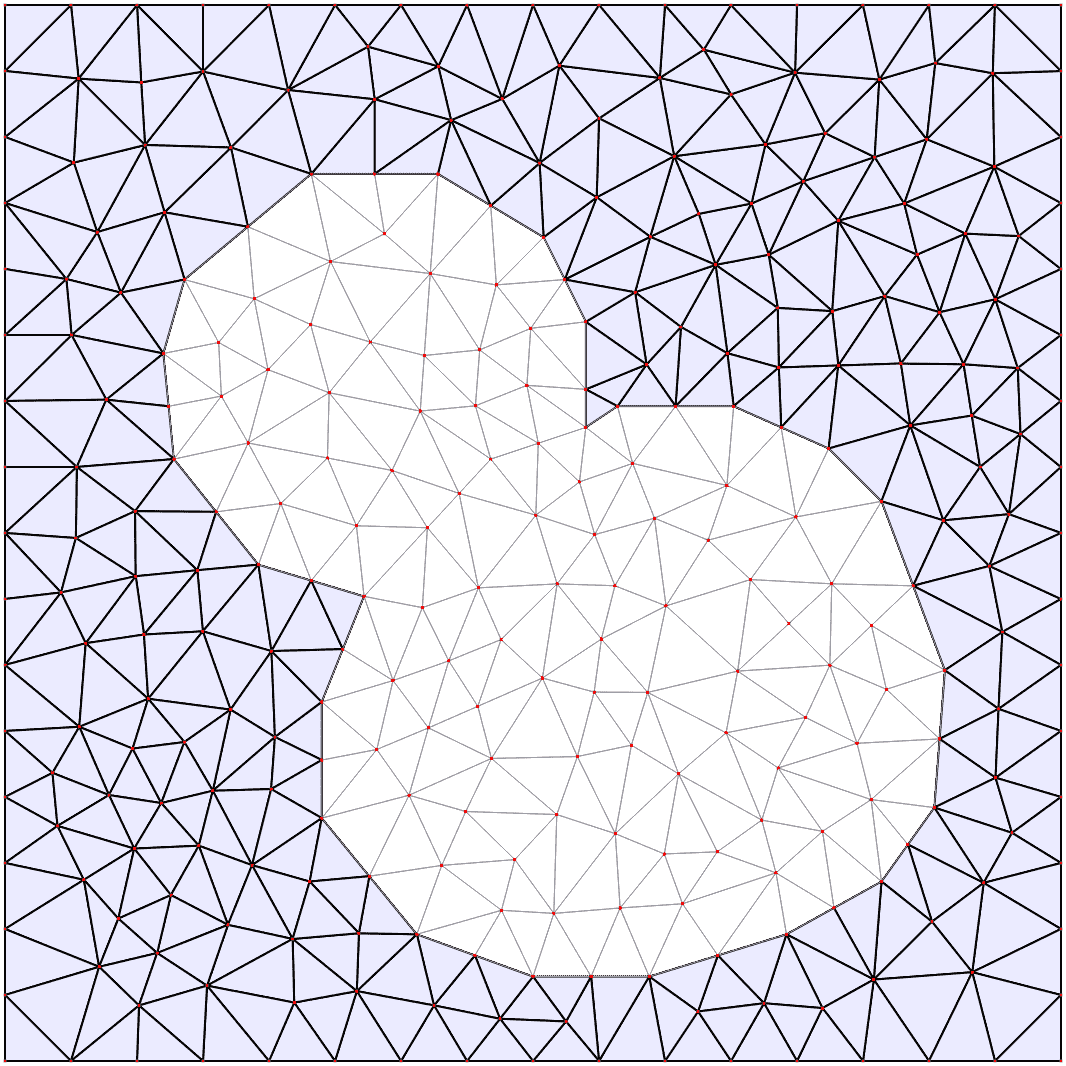
Automatic mesh generation
Starting from a segmented image of the compartments in the model, the boundaries are automatically identified and simplified, and from them a triangular mesh approximation to the geometry is constructed. For more details see the mesh documentation.
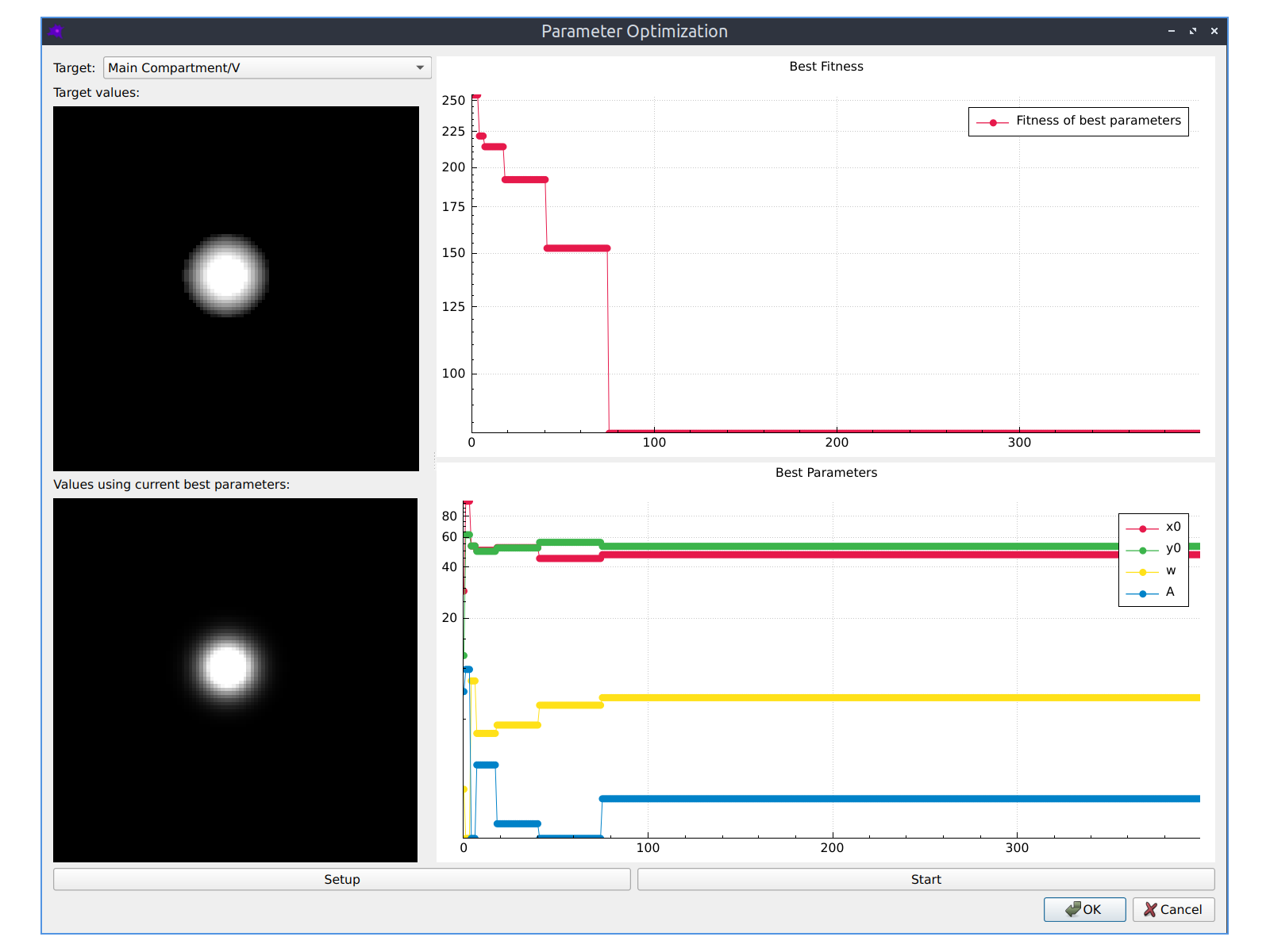
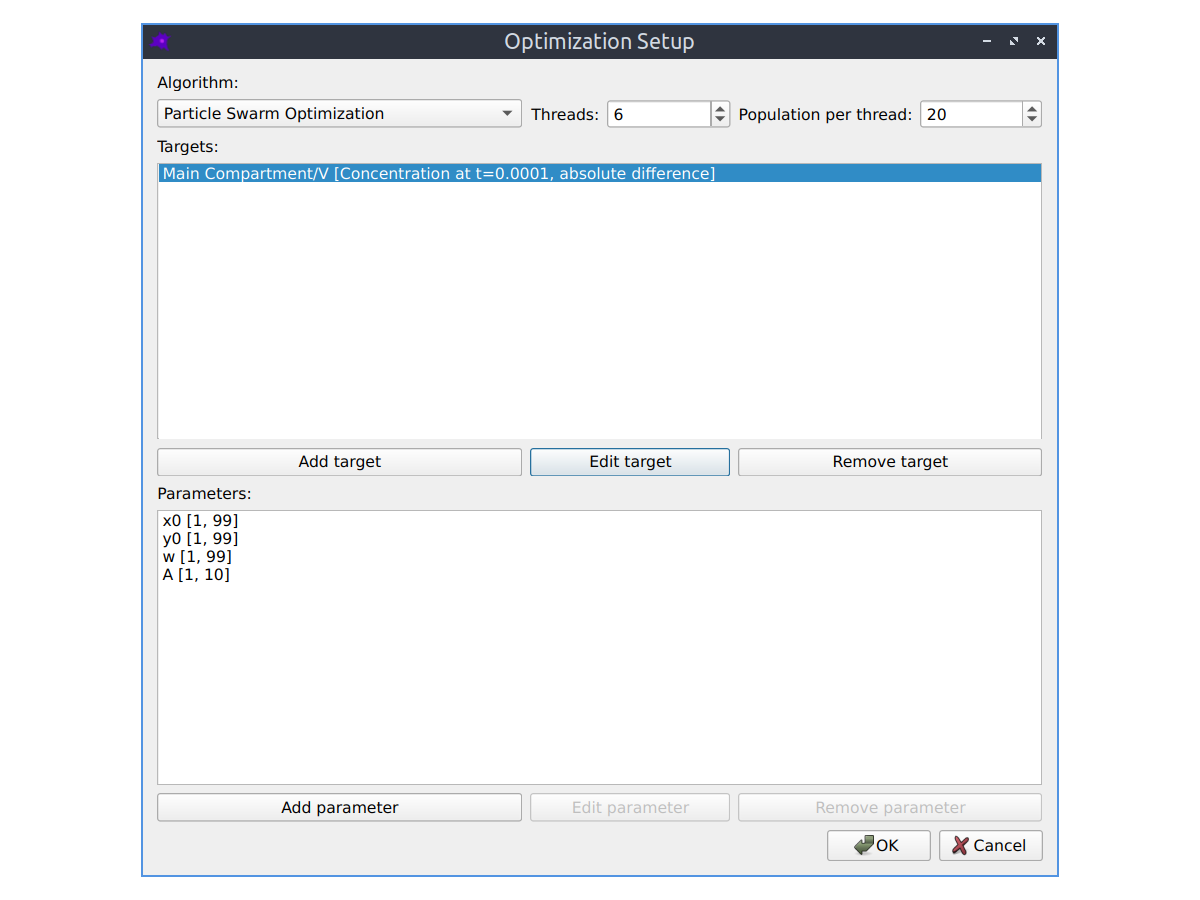
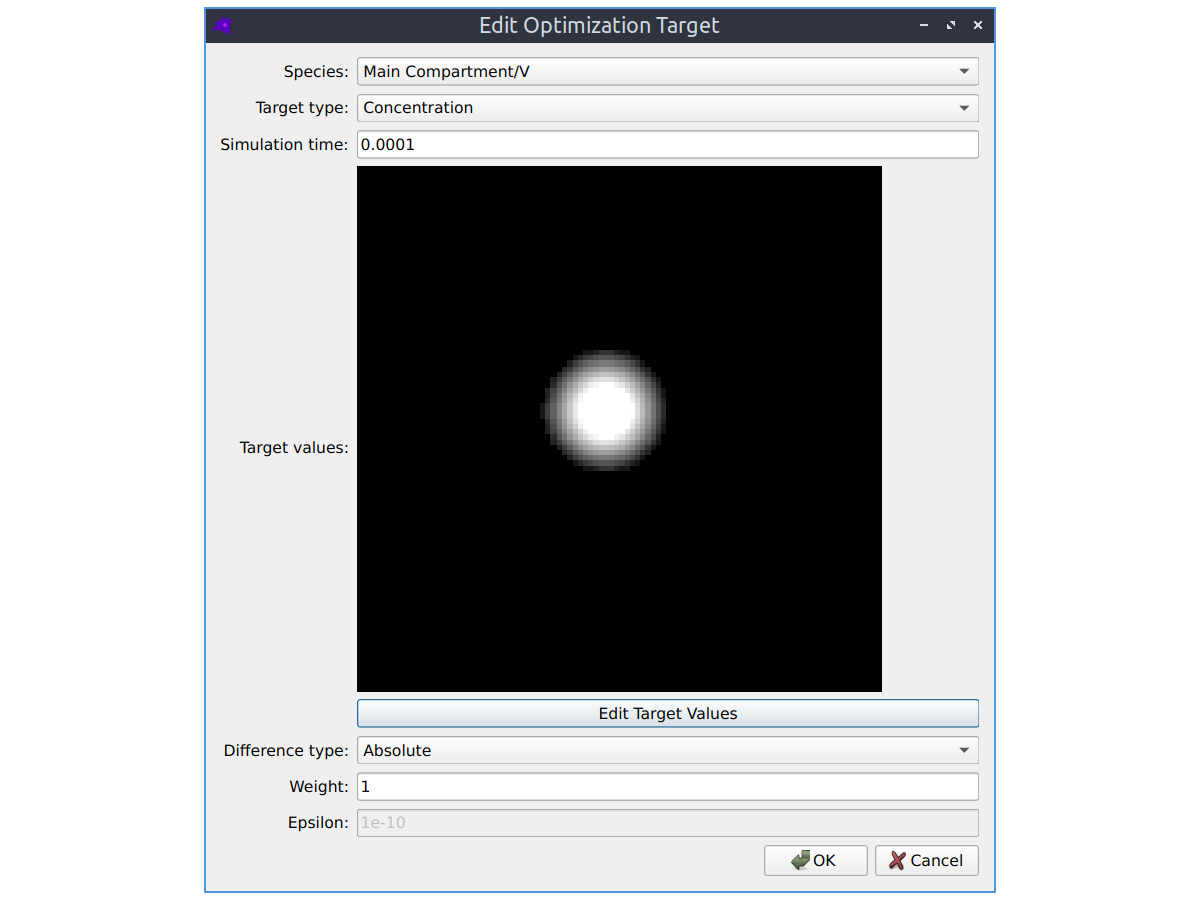
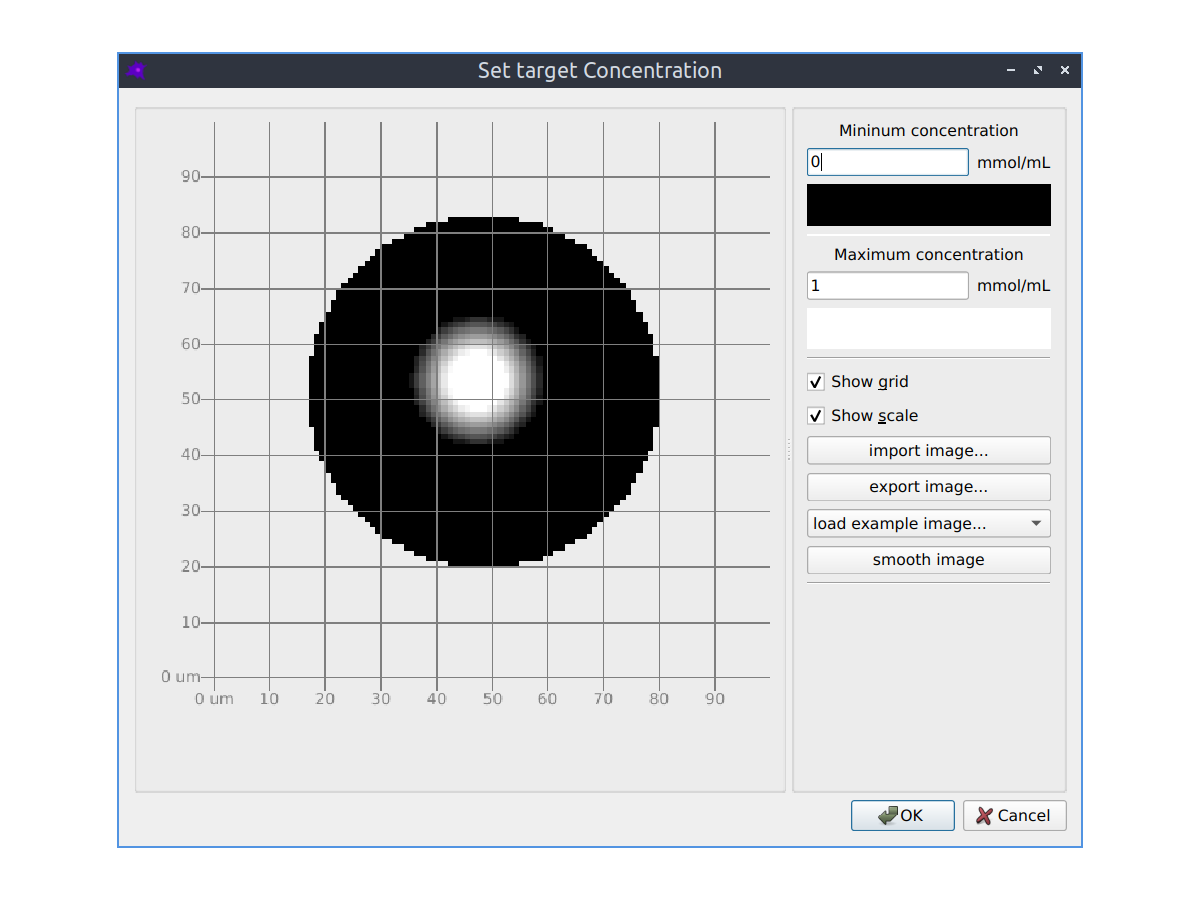
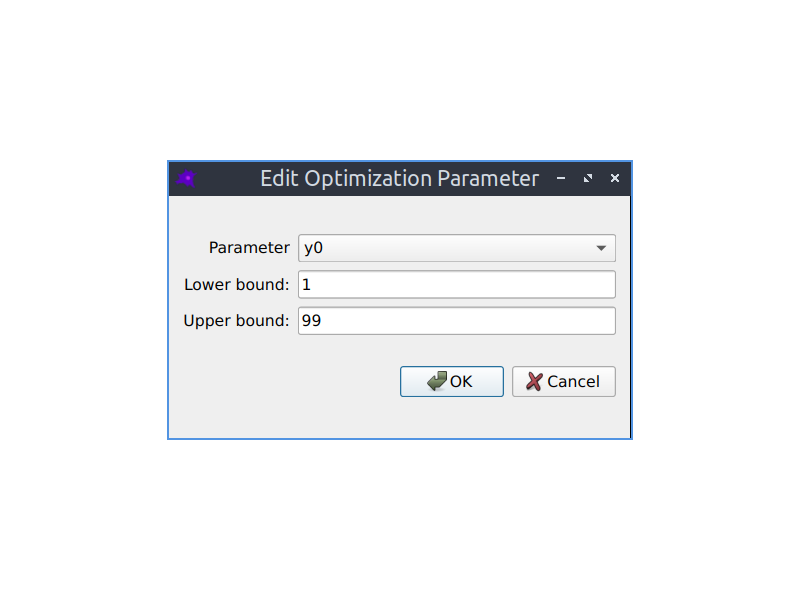
Parameter fitting
The parameters of your model can be automatically optimized using a variety of optimization algorithms to make the simulated model concentrations match experimental images of species concentrations. See the Parameter fitting section in the documentation to learn more.
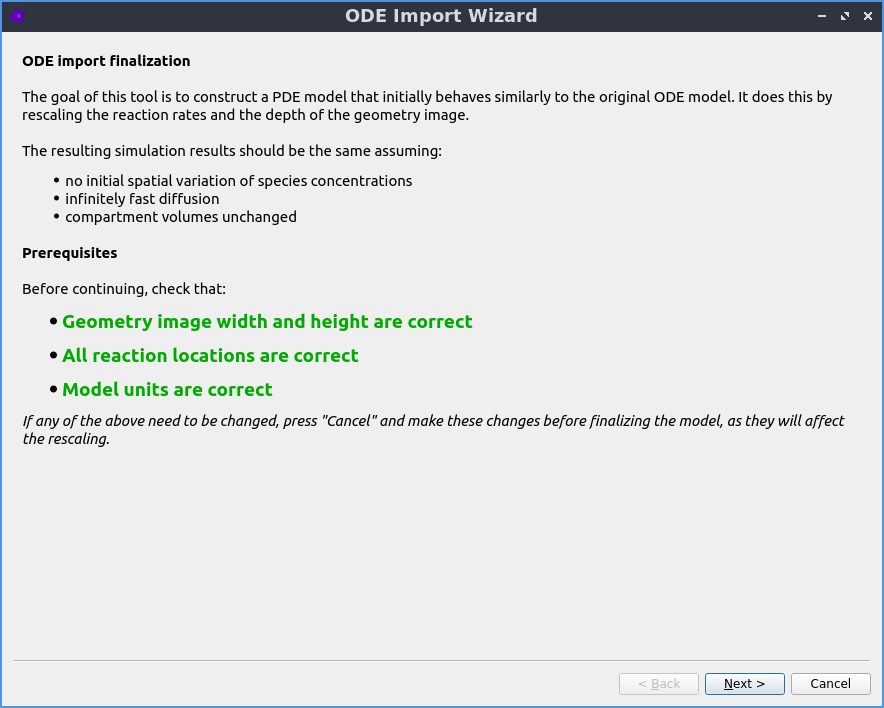
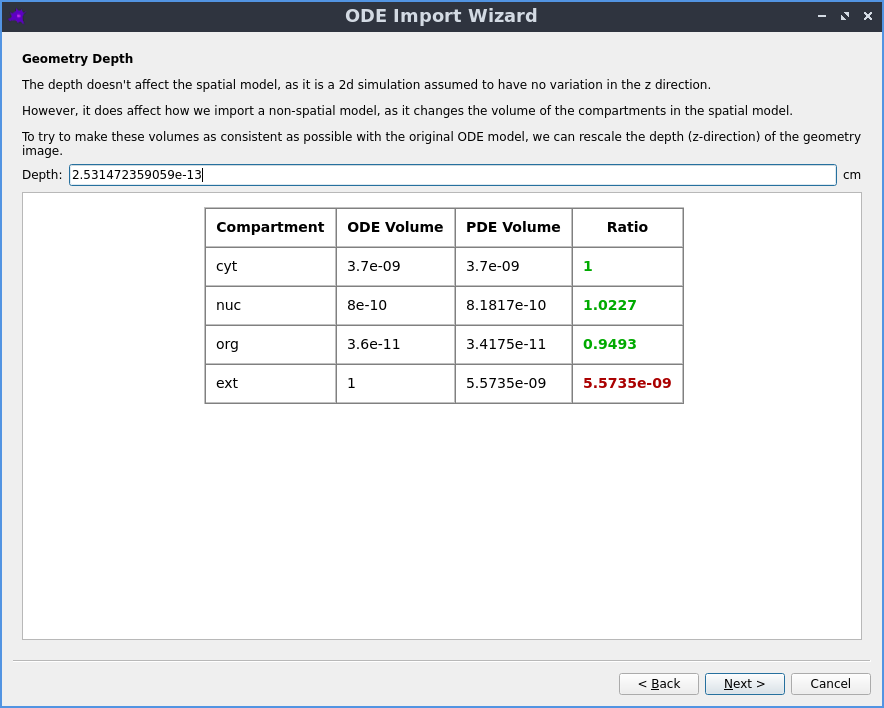
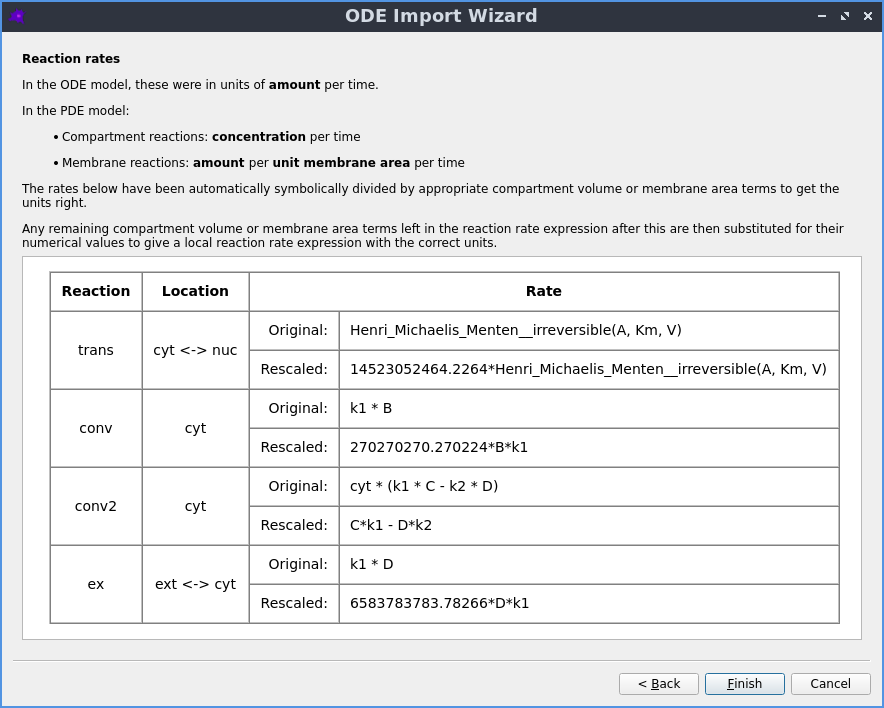
Automatic reaction rescaling
Guided workflow to import a non-spatial model from SBML xml or Copasi cps format, which automatically rescales the reaction rates to construct a valid spatial PDE model that initially reproduces the behaviour of the non-spatial ODE model.
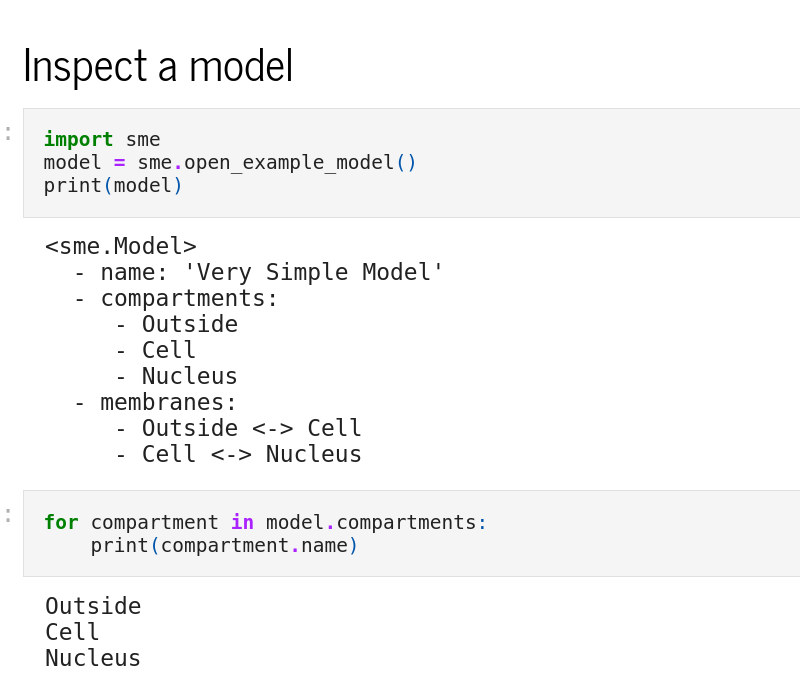
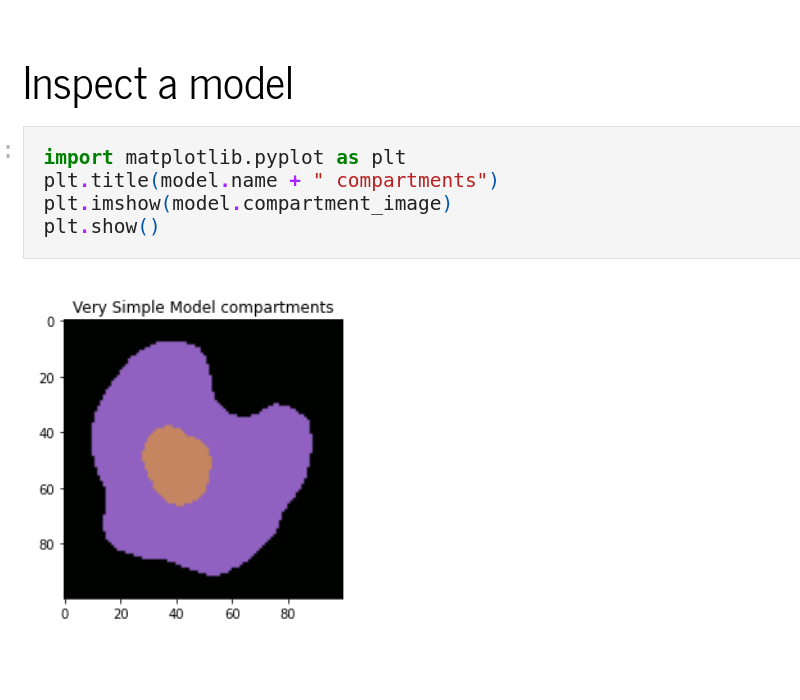
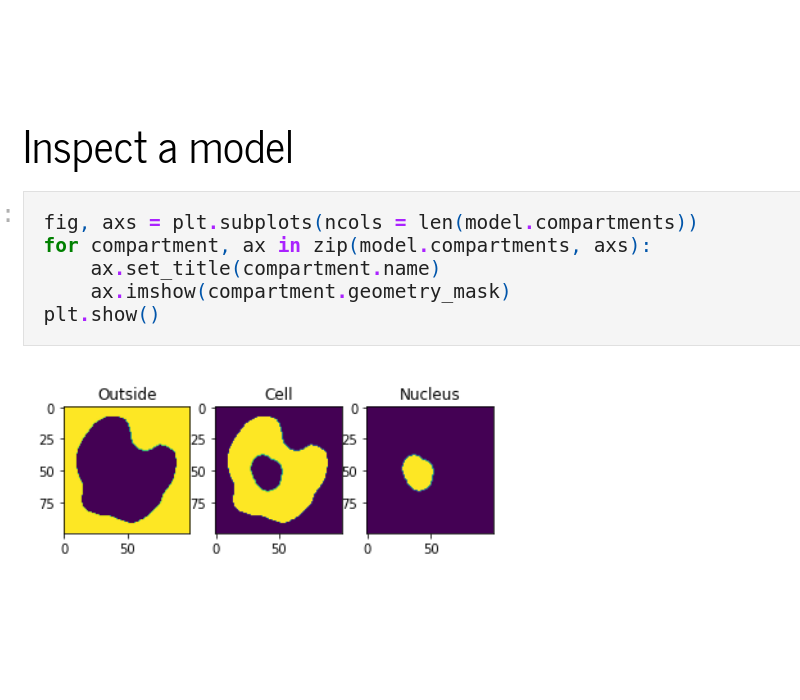
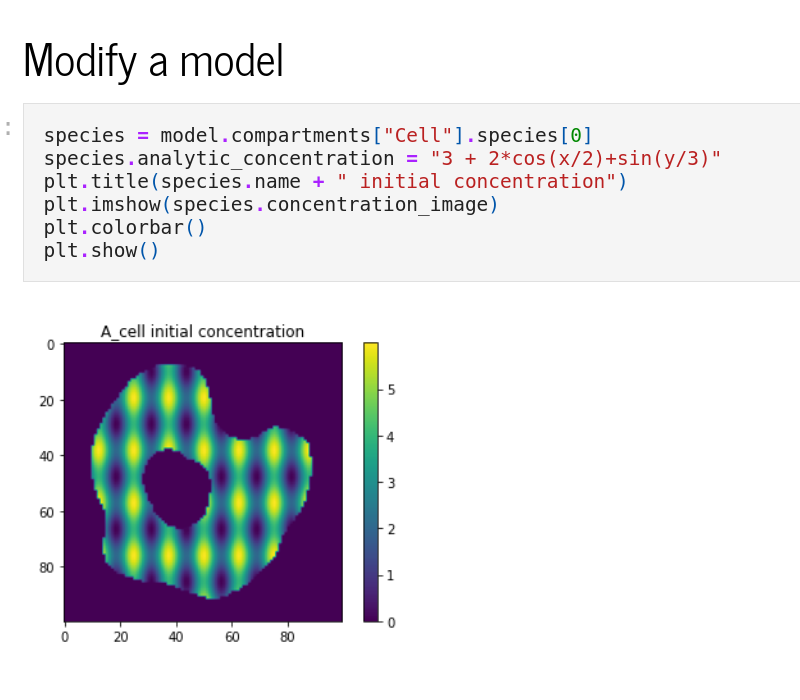
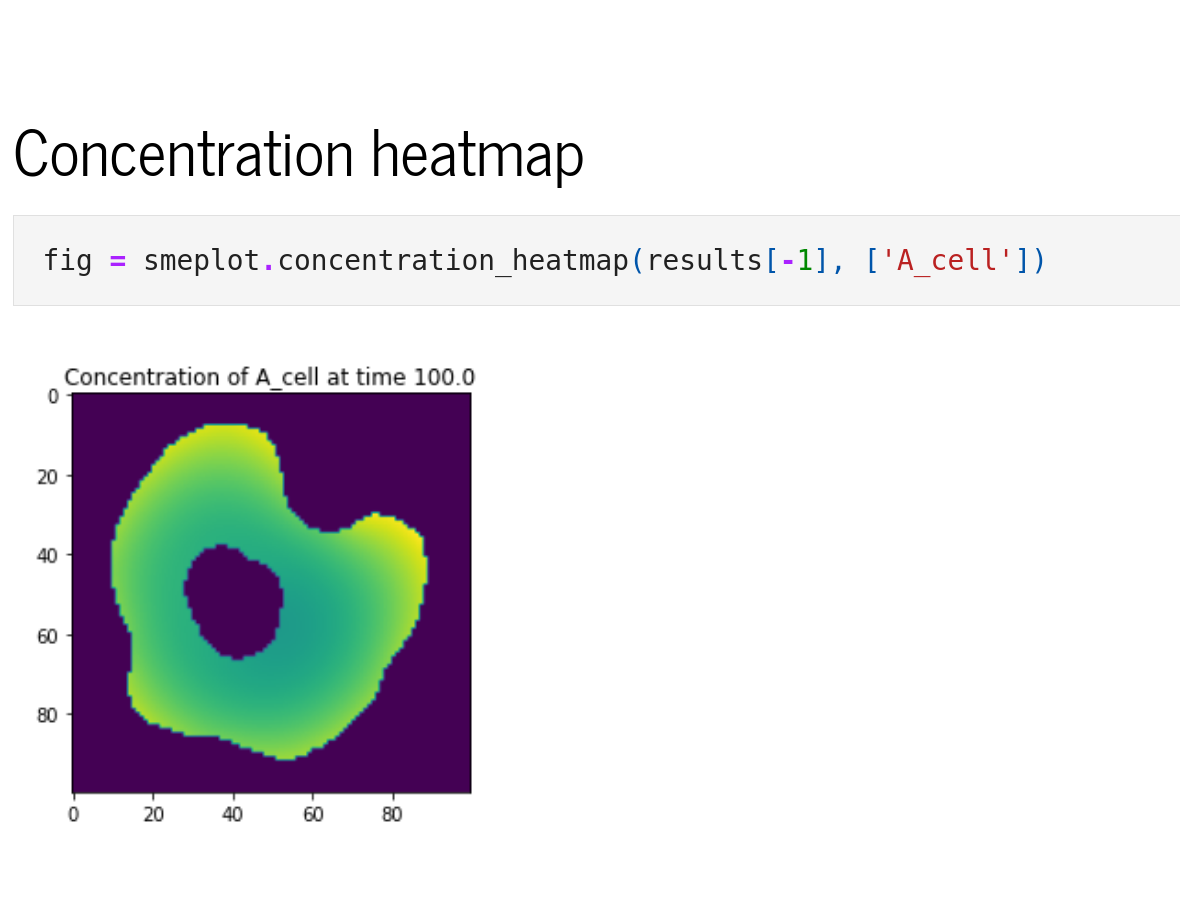
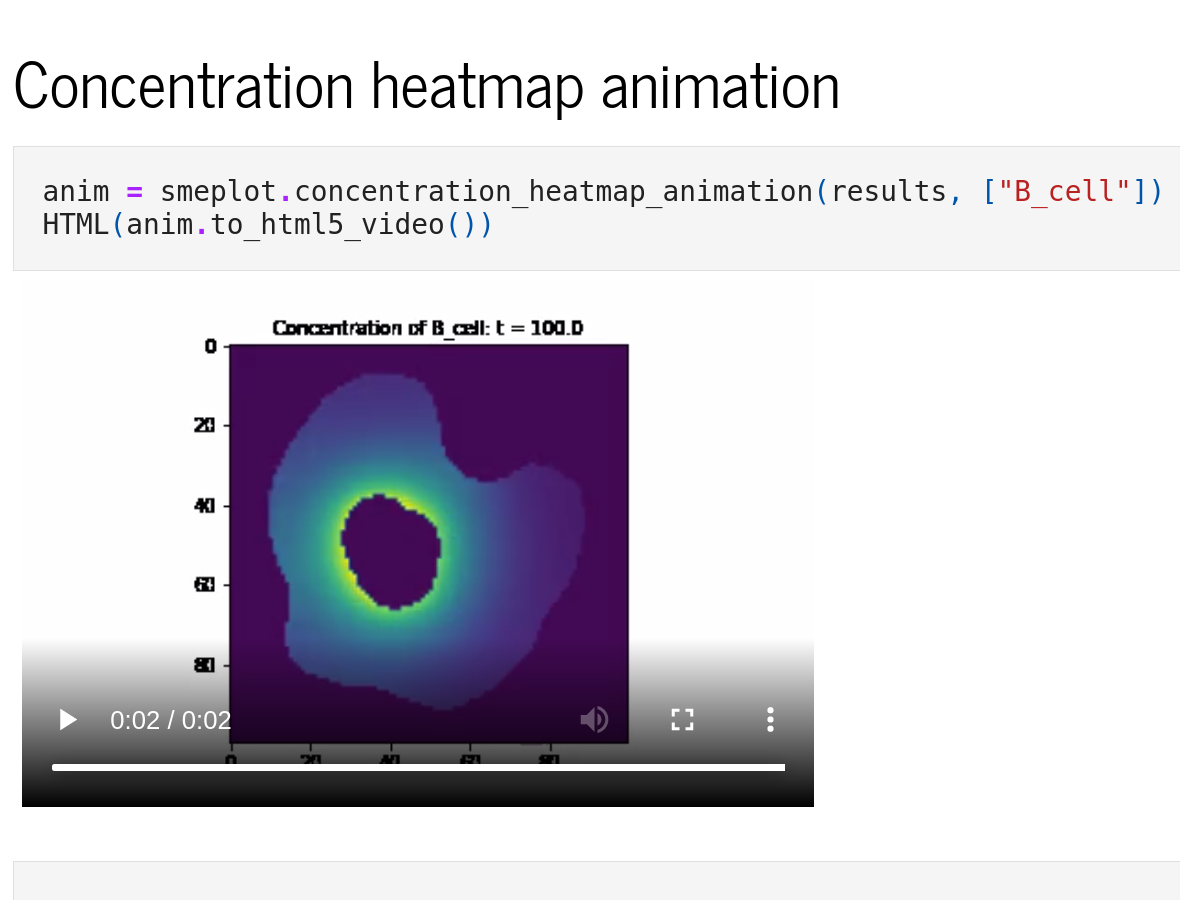
Python interface
The Python interface, sme, allows scripted editing and simulation of a model. See the Example Notebooks or the API reference for more information.
Python extras
Additional functionality such as plotting, animations and parameter fitting is provided by sme-contrib. See the documentation for more information and example notebooks.
SBML Spatial
Spatial models are imported and exported in SBML Level 3 format with the spatial package, using the libSBML library. All Level 3 math is supported, and is parsed, differentiated and compiled using the SymEngine symbolic manipulation library.